Checkpoints
Create Healthcare Dataset
/ 30
Set up IAM Permissions
/ 30
Create Data Stores
/ 20
Exporting metadata to BigQuery
/ 20
Ingesting DICOM Data with the Healthcare API
- GSP615
- Overview
- Healthcare API introduction
- Healthcare API concepts
- Setup and requirements
- Task 1. Create Healthcare dataset
- Task 2. Set up IAM permissions
- Task 3. Define variables needed
- Task 4. Create data stores
- Task 5. Import to DICOM datasets
- Task 6. Exporting metadata to BigQuery
- Task 7. Exploring the data in BigQuery
- Lab review
- Congratulations
- End your lab
GSP615
Overview
In this lab you will discover and use the basic functionality of Cloud Healthcare API using Digital Imaging and Communications in Medicine (DICOM) data model.
Healthcare API introduction
Cloud Healthcare API provides a managed solution for storing and accessing healthcare data in Google Cloud, providing a critical bridge between existing care systems and applications hosted on Google Cloud. Using the API, you can unlock significant new capabilities for data analysis, machine learning and application development, and use these capabilities to build the next generation of healthcare solutions.
The API is comprised of three modality-specific interfaces that implement key industry-wide standards for healthcare data:
- FHIR, an emerging standard for health data interchange
- HL7v2, the most widely adopted method for health systems integration
- DICOM, the dominant standard for radiology and imaging-related disciplines
Each interface is backed by a standards-compliant data store that provides read, write, search, and other operations on the data.
The Cloud Healthcare API provides a number of key features that are critical to bridging current technologies to the next generation of healthcare systems and applications:
- Standards conformance - Google supports the use of standards-based interoperability through its participation in a number of healthcare standards bodies. In the Cloud Healthcare API each modality-specific data store and its associated API is substantially conformant with its relevant standard. For example, FHIR stores implement STU3, the current version of the FHIR specification, and DICOM stores implement DICOMweb, a web-based standard for exchanging medical images. In future updates, we expect to support additional versions of these specifications as well as the ability to request a resource in a different version than its canonical representation.
- Compliance with privacy regulations - Google Cloud provides detailed guidance regarding how it supports compliance with HIPAA in the US, the PIPEDA in Canada, and other global privacy standards at cloud.google.com/security/compliance.
- Data location control - The Cloud Healthcare API treats data location as a core component of the API. You have the option to select the storage location for each dataset from a list of currently available locations which correspond to distinct geographic areas aligned with Google Cloud's regional structure. Future Google Cloud regions will allow for the distribution of storage across wider geographic areas.
- Security - The Cloud Healthcare API security model is based on Google's proven Identity and Access Management (IAM) system. IAM's fine-grained permissions give you complete control over access to your healthcare data. In addition, we've created open-source proxies for our powerful Apigee API Management system, which provides comprehensive threat detection and traffic management capabilities that allow you to securely expose sensitive ePHI with patient and provider applications.
- Bulk import and export - The Cloud Healthcare API's DICOM and FHIR modalities support bulk import and export of data, making it easier to transfer data via the Cloud Storage system.
- De-identification - De-identification support for DICOM is available, making it much easier to redact patient information from studies for research and other purposes. The de-identification process operates on a data store basis.
- Auditability - Both administrative and data access requests to the Cloud Healthcare API can be audited. Logs are available through Google Cloud's Stackdriver hybrid monitoring system.
- High availability - Availability for mission-critical scenarios is made possible through Google Cloud's robust and highly redundant infrastructure.
For many applications, the Cloud Healthcare API can provide a modern alternative to legacy stacks implementing DICOM, HL7v2 or FHIR STU3 standards, simplifying data integration with existing systems and enabling the application developers to focus on their differentiating features such as UX and intelligence.
In this lab, you will:
- Gain a general understanding of the Cloud Healthcare API and its role in managing healthcare data.
- Learn how to create Cloud Healthcare API datasets and stores..
- Import and Export DICOM data using the Cloud Healthcare API.
Healthcare API concepts
To get the most out of the Cloud Healthcare API, there are a few key concepts you'll want to understand. The information below should give you a good sense of Cloud Healthcare API capabilities, but you can find more details in the Cloud Healthcare API documentation.
General structure of the Cloud Healthcare API
The Cloud Healthcare API exposes interfaces that enable you to perform different types of functions:
- Administrative functions, such as creating or listing datasets and stores that will contain your data.
- Data access functions that allow you to create, update, delete and search the data stored in Cloud Healthcare API, or to perform bulk import and export operations.
- Security functions that allow you to impose access controls on data stored in Cloud Healthcare API.
- De-identification functions that allow you to replace ePHI with anonymized data, or to obfuscate ePHI so that it cannot be used.
- Metadata functions, such as retrieval of a FHIR capabilities statement for the FHIR API.
These functions may vary slightly depending on the modality of data (FHIR, HL7 v2 or DICOM) being operated on. For example, data retrieval operations against an FHIR data store use an API that conforms to the FHIR standard, but data retrieval operations against an HL7 v2 store use operations better suited to operating on HL7v2-structured data.
Datasets and stores
All Cloud Healthcare API usage occurs within the context of a Google Cloud project. Projects form the basis for creating, enabling, and using all Google Cloud services including managing APIs, enabling billing, adding and removing collaborators, and managing permissions for Google Cloud resources. Cloud Healthcare API can be used in one or many Google Cloud projects, as appropriate; this flexibility allows you to separate production from non-production usage, for example, or to segregate applications and resources in order to better manage access or accommodate different development lifecycles.
Within a project, data ingested through Cloud Healthcare API is stored in a dataset, which resides in a geographic location corresponding to a specific Google Cloud region. You use the Cloud Healthcare API's administrative functions to create a dataset in a particular location; doing so facilitates implementation of data location requirements for the countries in which your applications provide services. For example, you can choose to create a dataset in Google Cloud's "us-central1" region for US-based applications, or in an EU or UK region for applications serving those customers. This level of location control is also available in other Google Cloud products, which can be combined with Cloud Healthcare API to create a complete application architecture. A list of generally available Google Cloud products and the regions in which they are implemented can be found on Google Cloud, Cloud locations.
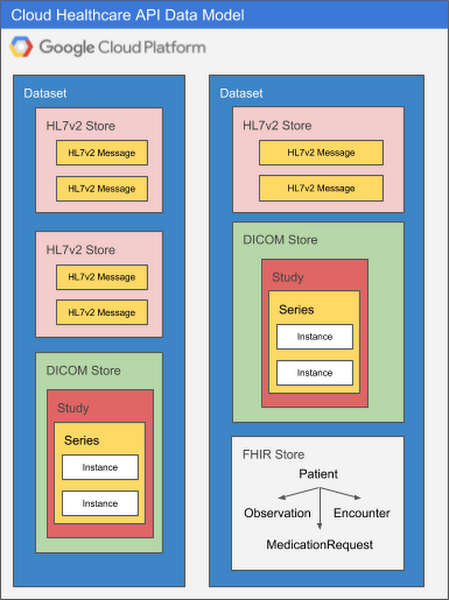
Because each healthcare data modality has different structural and processing characteristics, datasets are split into modality-specific stores. A single dataset can contain one or many stores, and those stores can all service the same modality or different modalities as application needs dictate. Using multiple stores in the same dataset might be appropriate if a given application processes different types of data, for example, or if you'd like to be able to separate data according to its source hospital, clinic, department, etc. An application can access as many datasets or stores as its requirements dictate with no performance penalty, so it's important to design your overall dataset and store architecture to meet the organization's broad goals for locality, partitioning, access control, and so on.
The diagram below illustrates two datasets in a Google Cloud project, each of which contains multiple stores.
There are many ways to structure datasets and stores. As you design systems that use the Cloud Healthcare API, you may want to take the following into consideration:
- Security and access control: Rules can be defined at both a dataset and store level, but you may choose to group all data for a particular application into the same dataset, and set access control rules such that only that application can access the dataset.
- Application requirements: An application processing different types of data may have all of its data for all modalities in a single dataset.
- Source systems: Often, the structure of healthcare data can vary according to the source system and modality. Separating data for different source systems into their own datasets may facilitate processing.
- Intended use: Data from different systems can have different intended uses, such as research, analytics, or machine learning predictions. Grouping data by intended use may facilitate ingestion into the target system.
- Separating ePHI from de-identified data: Cloud Healthcare API data de-identification functions read from a source dataset and write the output into a new dataset that you specify. If you are preparing data to be used by researchers, for example, this approach to de-identifying data may be a consideration in how you use datasets to segregate data.
MLLP Adapter
The minimal lower layer protocol (MLLP) is the standard used for transmitting HL7v2 messages over TCP/IP connections within a network, such as a hospital.
MLLP does not offer an exact mapping to the Cloud Healthcare API HL7v2 REST API], which uses HTTP. Therefore, an MLLP adapter must be used to convert messages transmitted over MLLP into a format that an HTTP/REST API can accept. To transmit messages over MLLP and then to the Cloud Healthcare API, use the Google Cloud MLLP adapter.
There are many ways to structure datasets and stores. As you design systems that use the Cloud Healthcare API, you may want to take the following into consideration:
- Security and access control: Rules can be defined at both a dataset and store level, but you may choose to group all data for a particular application into the same dataset, and set access control rules such that only that application can access the dataset.
- Application requirements: An application processing different types of data may have all of its data for all modalities in a single dataset.
- Source systems: Often, the structure of healthcare data can vary according to the source system and modality. Separating data for different source systems into their own datasets may facilitate processing.
- Intended use: Data from different systems can have different intended uses, such as research, analytics or machine learning predictions. Grouping data by intended use may facilitate ingestion into the target system.
- Separating ePHI from de-identified data: Cloud Healthcare API data de-identification functions read from a source dataset and write the output into a new dataset that you specify. If you are preparing data to be used by researchers, for example, this approach to de-identifying data may be a consideration in how you use datasets to segregate data.
Setup and requirements
Before you click the Start Lab button
Read these instructions. Labs are timed and you cannot pause them. The timer, which starts when you click Start Lab, shows how long Google Cloud resources will be made available to you.
This hands-on lab lets you do the lab activities yourself in a real cloud environment, not in a simulation or demo environment. It does so by giving you new, temporary credentials that you use to sign in and access Google Cloud for the duration of the lab.
To complete this lab, you need:
- Access to a standard internet browser (Chrome browser recommended).
- Time to complete the lab---remember, once you start, you cannot pause a lab.
How to start your lab and sign in to the Google Cloud console
-
Click the Start Lab button. If you need to pay for the lab, a pop-up opens for you to select your payment method. On the left is the Lab Details panel with the following:
- The Open Google Cloud console button
- Time remaining
- The temporary credentials that you must use for this lab
- Other information, if needed, to step through this lab
-
Click Open Google Cloud console (or right-click and select Open Link in Incognito Window if you are running the Chrome browser).
The lab spins up resources, and then opens another tab that shows the Sign in page.
Tip: Arrange the tabs in separate windows, side-by-side.
Note: If you see the Choose an account dialog, click Use Another Account. -
If necessary, copy the Username below and paste it into the Sign in dialog.
{{{user_0.username | "Username"}}} You can also find the Username in the Lab Details panel.
-
Click Next.
-
Copy the Password below and paste it into the Welcome dialog.
{{{user_0.password | "Password"}}} You can also find the Password in the Lab Details panel.
-
Click Next.
Important: You must use the credentials the lab provides you. Do not use your Google Cloud account credentials. Note: Using your own Google Cloud account for this lab may incur extra charges. -
Click through the subsequent pages:
- Accept the terms and conditions.
- Do not add recovery options or two-factor authentication (because this is a temporary account).
- Do not sign up for free trials.
After a few moments, the Google Cloud console opens in this tab.
Activate Cloud Shell
Cloud Shell is a virtual machine that is loaded with development tools. It offers a persistent 5GB home directory and runs on the Google Cloud. Cloud Shell provides command-line access to your Google Cloud resources.
- Click Activate Cloud Shell
at the top of the Google Cloud console.
When you are connected, you are already authenticated, and the project is set to your Project_ID,
gcloud is the command-line tool for Google Cloud. It comes pre-installed on Cloud Shell and supports tab-completion.
- (Optional) You can list the active account name with this command:
- Click Authorize.
Output:
- (Optional) You can list the project ID with this command:
Output:
gcloud, in Google Cloud, refer to the gcloud CLI overview guide.
Task 1. Create Healthcare dataset
- From the Console, go to Navigation menu > Healthcare.
-
ENABLE the API.
-
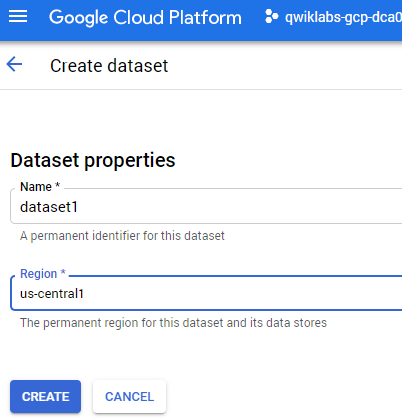
Now click +CREATE DATASET.
-
Name the dataset "dataset1" and select the
region.
- Then click Create.
Click Check my progress to verify the objective.
Task 2. Set up IAM permissions
- Edit the permissions for your Cloud Healthcare Service Agent by running the following command in Cloud Shell:
Click Check my progress to verify the objective.
Task 3. Define variables needed
- In Cloud Shell run the following to set variables needed for this lab:
Task 4. Create data stores
Data in Cloud Healthcare API datasets and stores can be accessed and managed using a REST API that identifies each store using its project, location, dataset, store type and store name. This API implements modality-specific standards for access that are consistent with industry standards for that modality. For example, the Cloud Healthcare DICOM API natively provides operations for reading DICOM studies and series that are consistent with the DICOMweb standard, and supports the DICOM DIMSE C-STORE protocol via an open-source adapter.
- Call the API to create a DICOM store:
The server returns a path to the newly created store.
Click Check my progress to verify the objective.
You can also use the curl utility to issue Cloud Healthcare API calls. curl is pre-installed in your Cloud Shell machine. By default, curl does not show HTTP status codes or session-related information; if you would like to see this information please add the "-v" option to all commands in this tutorial.
- Try creating a secondary DICOM store by using the below command:
Operations that access a modality-specific store use a request path that is comprised of two pieces: a base path, and a modality-specific request path.
Administrative operations — which generally operate only on locations, datasets and stores — may only use the base path, but data modality-specific retrieval operations use both the base path (for identifying the store to be accessed) and request path (for identifying the actual data to be retrieved).
Task 5. Import to DICOM datasets
In this section you will be importing data from the NIH Chest x-ray set to a DICOM store. For more information on the public dataset, visit the Public datasets documentation.
- Call the API to use the import functionality:
- Continue to issue this option status request until the result shows
donein the response.
Task 6. Exporting metadata to BigQuery
- Run the following to create a dataset in BigQuery:
- Export the metadata to a new table:
Click Check my progress to verify the objective.
Task 7. Exploring the data in BigQuery
-
In the Console, navigate to Navigation menu > BigQuery.
-
Under Explorer find your project ID and expand the drop-down.
-
Find dataset1 and expand the drop-down.
-
Select dicomstore1 under the drop down and navigate the the Preview tab, where the recent imported metadata is displayed. Scroll right to see the data that was imported into the table.
-
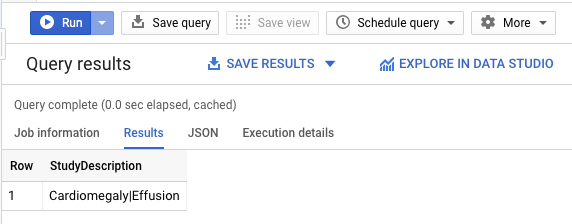
In the +Create SQL query add the following query, then click RUN to find out what patient 25290's findings are:
See that a Effusions with Cardiomegaly was found.
- Click on +Create SQL query and in the query EDITOR now add the following query to find out who the patients with no findings are:
Lab review
Cloud Healthcare API provides a comprehensive facility for ingesting, storing, managing, and securely exposing healthcare data in FHIR, DICOM, and HL7 v2 formats. Using Cloud Healthcare API, you can ingest and store data from electronic health records systems (EHRs), radiological information systems (RISs), and custom healthcare applications. You can then immediately make that data available to applications for analysis, machine learning prediction and inference, and consumer access.
Cloud Healthcare API enables application access to healthcare data via widely-accepted, standards-based interfaces such as FHIR STU3 and DICOMweb. These APIs allow data ingestion into modality-specific data stores, which support data retrieval, update, search and other functions using familiar standards-based interfaces.
Further, the API integrates with other capabilities in Google Cloud through two primary mechanisms:
- Cloud Pub/Sub, which provides near-real-time updates when data is ingested into a Cloud Healthcare API data store, and
- Import/export APIs, which allow you to integrate Cloud Healthcare API into both Google Cloud Storage and Google BigQuery.
Using Cloud Pub/Sub with Google Cloud Functions enables you to invoke machine learning models on healthcare data, storing the resulting predictions back in Cloud Healthcare API data store. A similar integration with Cloud Dataflow supports transformation and cleansing of healthcare data prior to use by applications.
To support healthcare research, Cloud Healthcare API offers de-identification capabilities for FHIR and DICOM. This feature allows customers to share data with researchers working on new cutting-edge diagnostics and medicines.
Congratulations
In this lab you:
- Gained a general understanding of Cloud Healthcare API and its role in managing healthcare data.
- Learned how to create datasets and stores for FHIR and DICOM data.
- Imported FHIR and DICOM data.
Finish your quest
This self-paced lab is part of the Cloud Healthcare API quest. A quest is a series of related labs that form a learning path. Completing this quest earns you a badge to recognize your achievement. You can make your badge or badges public and link to them in your online resume or social media account. Enroll in this quest or any quest that contains this lab and get immediate completion credit. See the Google Cloud Skills Boost catalog to see all available quests.
Take your next lab
Continue your quest with Ingesting DICOM Data with the Healthcare API or try one of these suggestions:
End your lab
When you have completed your lab, click End Lab. Your account and the resources you've used are removed from the lab platform.
You will be given an opportunity to rate the lab experience. Select the applicable number of stars, type a comment, and then click Submit.
The number of stars indicates the following:
- 1 star = Very dissatisfied
- 2 stars = Dissatisfied
- 3 stars = Neutral
- 4 stars = Satisfied
- 5 stars = Very satisfied
You can close the dialog box if you don't want to provide feedback.
For feedback, suggestions, or corrections, please use the Support tab.
Manual last updated May 14, 2024
Lab last tested May 14, 2024
Copyright 2024 Google LLC All rights reserved. Google and the Google logo are trademarks of Google LLC. All other company and product names may be trademarks of the respective companies with which they are associated.